Structure and Hierarchy¶
The flowchart below is a simplified class diagram to show how the major objects in NutMEG interact with each other for a simple microbial growth simulation.
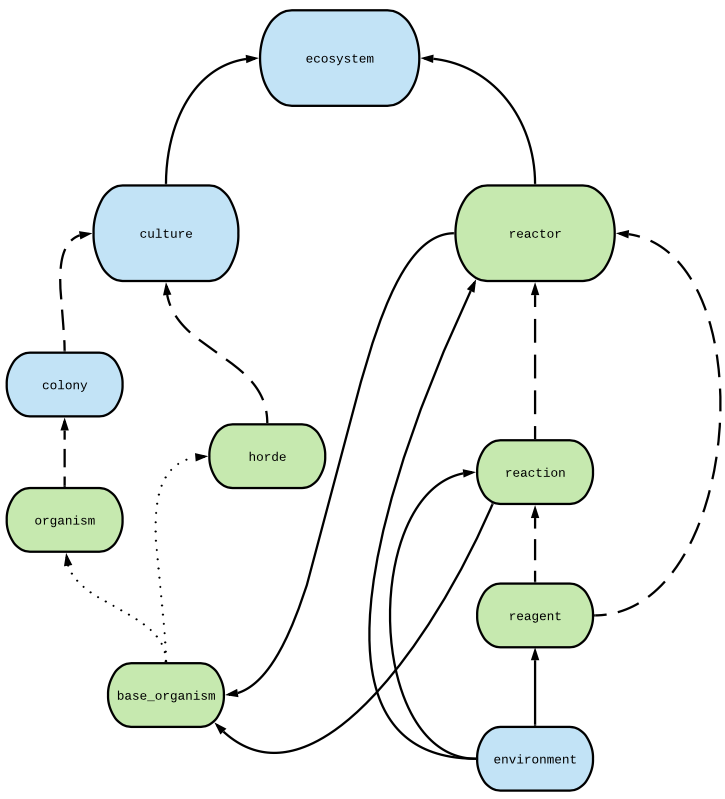
Basic hierarchy in layman’s terms¶
environment¶
The lowest level class contains global standard thermodynamic parameters such as the pressure, temperature and pressure of a system, as well as some other handy variables such as the volume the system would have at RTP.
reagent¶
Reagent objects store properties of chemical species in a given environment. There is not one reagent per molecule, rather this submodule characterises the whole collection of a single species, allowing us to know/calculate concentrations, activities, and thermodynamic data. Thermodynamic data at a range of temperatures and pressures (the ones stored in environment) can be calculated from chemical databases.
reaction¶
Reaction objects model chemical reactions by taking in a reaction equation and a collection of reagent s in the same environment. Thermodynamic considerations for various reaction types are included so far, such as purely thermodynamic (default), redox at nonstandard conditions, the full dissolution of salts, and ionic strength/activity calculators for ions in solution (in the reaction.special.solutions module). Further inclusions will probably come in the future as we explore new pathways. For each of these scenarios, reaction can perform a reaction — hence altering the properties of the reagent s — and calculate free energy yield per mole, amongst other things.
reactor¶
collects together all the reaction s of interest in a system along with all of the relevant reagent s, unifying them in a single environment. By default, reactor only works in well-mixed near-equilibrium conditions where the composition does not appreciably change unless we introduce something to disrupt the system such as an organism. e.g. there is no active chemistry element to it. Some kind of environmental maintained disequilibrium could be introduced, such as a constant fresh supply of a reagent.
Organism options¶
Option 1: base_organism and colony (computationally expensive)¶
A base_organism object describes an individual simple organism attempting to survive in your reactor. It has several parameters such as the obvious — mass, volume etc. — and some less intuitive such as death rates and maximum metabolic rates. There are other NutMEG-specific objects which base_organisms use, respirator, maintainer, CHNOPSexchanger`, and cell_synthesis as well as the adaptations submodule. All of these help compute various aspects of the organism’s chance of survival. base_organism also has the crucial take_step method, where the metabolism is performed with the reactor and survival/growth is predicted.
If you want to model each organism individually, they can be placed into a colony by species. A colony is simply a collection of base_organism-like objects to ease numerical modelling. The issue with simulating colony s is that when you have a large number of organisms they can become very computationally expensive very quickly, so this method is best left for smaller or extremely energy/nutrient limited systems. In most cases, it would be best to use a horde.
Option 2: horde (preferred)¶
A horde object is a child class of base_organism and essentially combines the two classes above. This significantly improves computation speed but you have less fidelity when considering individual organisms, for example if you wanted to monitor minor discrepancy / directed evolution between them.
culture¶
An object to collect together all organism behaviour in NutMEG. A culture can contain one or more hordes and colonies, each of those based on (or including) a
base_organism. Each horde/organism requires their own maintainer, respirator and CHNOPSexchanger object (or they can just use the default).
ecosystem¶
Object for performing simulations with the above. Fundamentally an ecosystem takes a culture and a reactor and performs/monitors how they interact. ecosystem s are capable of performing standard growth curve predictions.
applications¶
applications is a module full of specific classes designed to look at more than simple growth curves. Check out its API to see what else you can do!